In March, we announced our lead investment in the Series A for Pinecone, a semantic search database powered by vector embedding search. I firmly believe that the next decade of enterprise applications will use semantic search technology, whether it’s security, observability, or e-commerce. Any application would be immediately upgraded by moving from text search to semantic search. What’s significant is that historically tech like this was only accessible to hyper-scale companies such as the FANG’s. With Pinecone, the technology is available in the cloud and entirely self-serve.
Following that investment, I announced that I had open-sourced a Julia library that performs read/write/query/delete (CRUD) operations on the Pinecone. However, I didn’t show you how to use it — I’m going to change that here with a fascinating use case. Also, I’m sure this is much more interesting than a simple funding announcement article where you learn more about their financing and how Pinecone works. There are tons of that here and here, but I’ll follow up with one from the Menlo perspective soon.
In this post, I will show how to answer an important question that all parents have about their kids: “Which parent looks like their children most?”
To achieve this, I wrote a Julia program (with Python inlined using PyCall) to use the Julia->Pinecone library. The steps we’ll follow are the following:
- Get around three dozen photos each of myself, wife, and two younger kids
- Use DeepFace and grab embeddings from all of the images
- Upload those vector embeddings to a Pinecone index named “kids-<model>” [tested every model provided by DeepFace]
- Query the vectors representing the kids using vector data from myself and my wife.
- Check whether my wife or I are closer in vector distance (more similar).
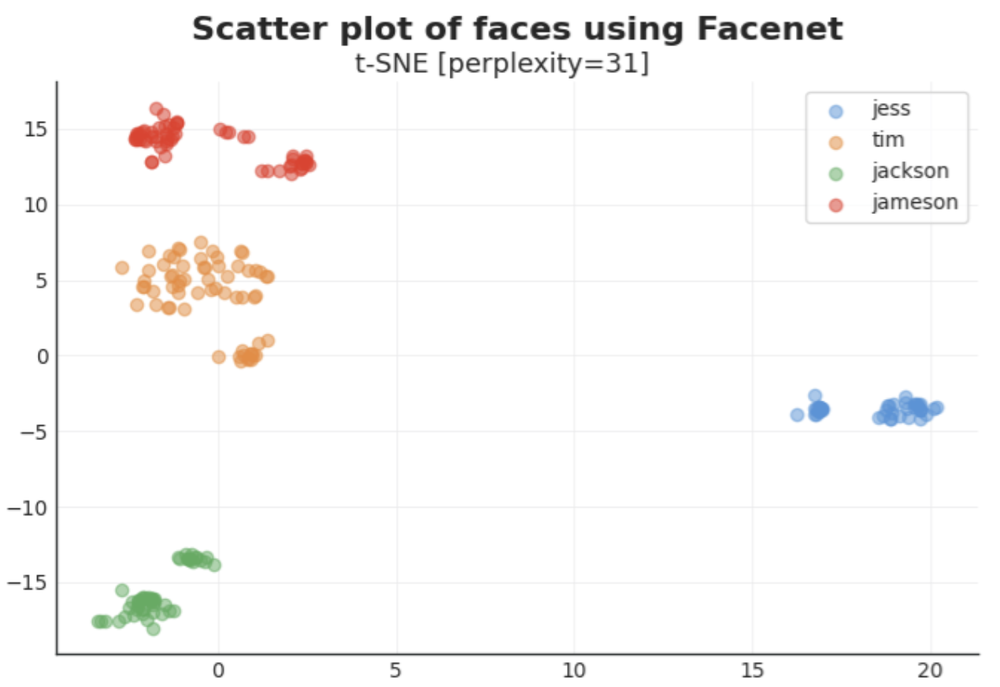
- Reduce the dense vectors once using PCA and then again w/ t-SNE for a pretty scatter plot
To explore how this works, let’s break down each step.
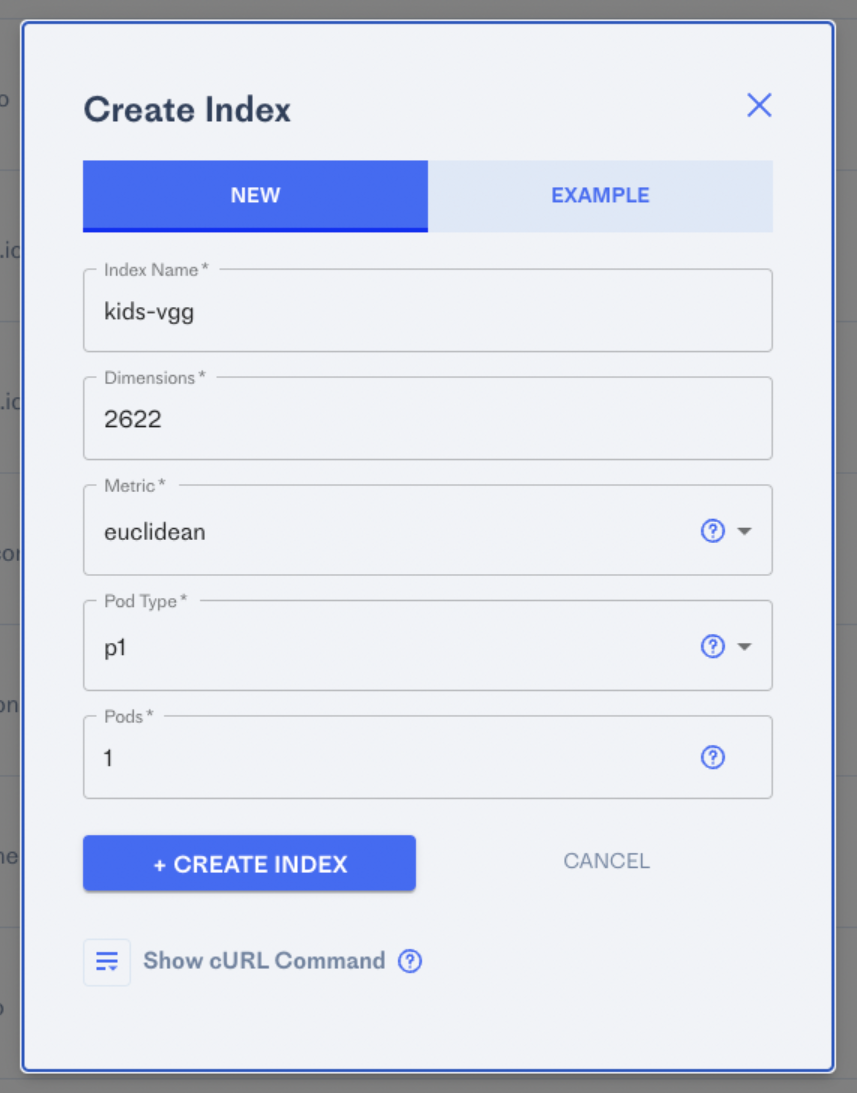
- Create a vector/embedding index in Pinecone. This is super easy as Pinecone is entirely self-serve, including payment integration w/ Stripe. Setting up an index takes seconds. For this example, we’ll use euclidean distance for measuring similarity. The “width” of the table is the number of elements or dimensions in your vectors. This is an example of trying out VGG from within DeepFace, which has a vector length of 2,622:
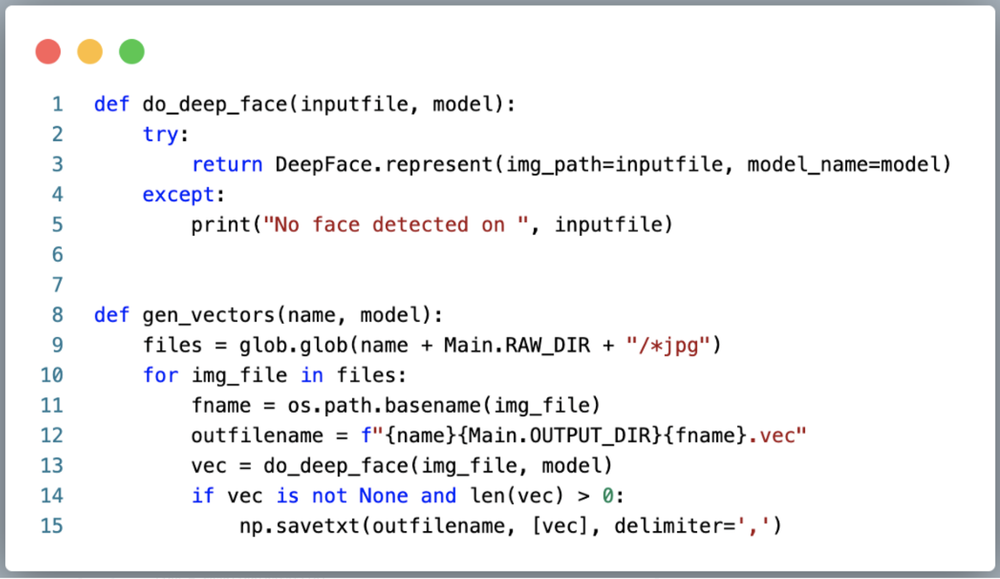
2. Vectorize the data using DeepFace. Below, we are simply globbing all of the face images generated above, feeding them into DeepFace, and saving the embeddings CSV style (one vector file per face image.)
3. Upload vectors to Pinecone. Now we are switching back to Julia. Note that this was all seamless, all in the same Julia file. The Python invoked from Julia above was in the same .jl file, but wrapped using PyCall with just py”””
Here in the code below, we are simply globbing all of the vector files generated above, reading them in, and uploading them to Pinecone. Note that Pinecone has an excellent metadata feature to tag each vector. In our example, we tag each vector with the first name of the person the vector belongs from. Metadata is simply a dictionary of key/value pairs. Here, I’m storing the persons first name with each vector in a Julia Dict.

Note we finally get to use the Julia=>Pinecone API with the Pinecone.upsert(…) call on line 12.
4. Query for Results. Finally, we get the most exciting part — which parent looks like each child the most!? The below is a simple function that takes a parent/child pair as args, reads in the parent vector from local disk, and uses that to query Pinecone for euclidean distance. Pinecone returns a JSON object that we parse using Julia’s JSON3, grab matches and sort them, then show the top two. Note the usage of the metadata feature in Pinecone to filter out only the vectors for a given child (line 2) with that Dict passed to the query (line 9).
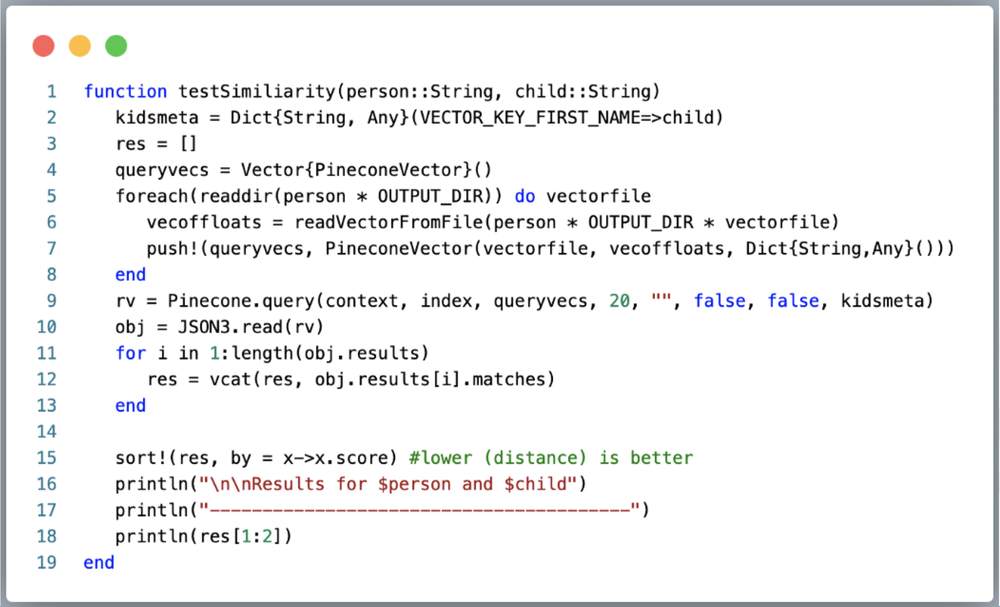
We just called testSimilarity(parent, child) for all combinations.

The results of querying Pinecone on vectors are shown below (smaller is closer in similarity). Sadly as suspected by both Jess and me, both of the kids are more similar to her. Note that for kicks, I also queried similarity between the two of us, and we are less similar to each other than we are to either of the kids. Note that DeepFace provides an abstraction for [“VGG-Face”, “Facenet”, “Facenet512”, “OpenFace”, “DeepFace”, “DeepID”, “ArcFace”, “Dlib”, “SFace”], and no matter which model I used, the results were always the same — Jess was more similar.

5. Generate plots. t-SNE is popular here for looking at clustering of images. Here we read in each vector file, make a first dimension reduction using PCA to 20 dims, then further reduce to two using TSNE and return that as a 2 column DataFrame for plotting. Then really, it’s a bunch of customization as Matplotlib/Seaborn default graphs are mostly hideous.
Aside from a nice visual, we can see that there is clear similarity/clustering for each person, which, more than anything, shows that images belonged to the right people. Note that distance between clusters is not indicative of similarity, a seemingly common mistake w/ t-SNE plots.

The source code shown above is available in the GitHub repo for this project.
Conclusion
So what are the key takeaways?
- The Julia API for Pinecone works great!
- The hypothesis that the kids look more similar to Jess has been confirmed with data.
- No matter which model was abstracted away by DeepFace (VGG-Face, Facenet, Dlib, etc) the results were always the same with respect to vector distance.
- Pinecone is incredibly powerful technology that provides a cloud vector embedding database for any number of semantic search use cases. Get started and learn more here!
Tim is a partner at Menlo Ventures focused on early-stage investments that speak to his passion for AI/ML, the new data stack, and the next-generation cloud. His background as a technology builder, buyer, and seller informs his investments in companies like Pinecone, Neon, Edge Delta, JuliaHub, TruEra, ClearVector, Squint, and…







